PAST: a Prior-based self-Attention method for Spatial Transcriptomics¶
News¶
Souce code available!¶
PAST is now available on GitHub 2022-10-14
Overview¶
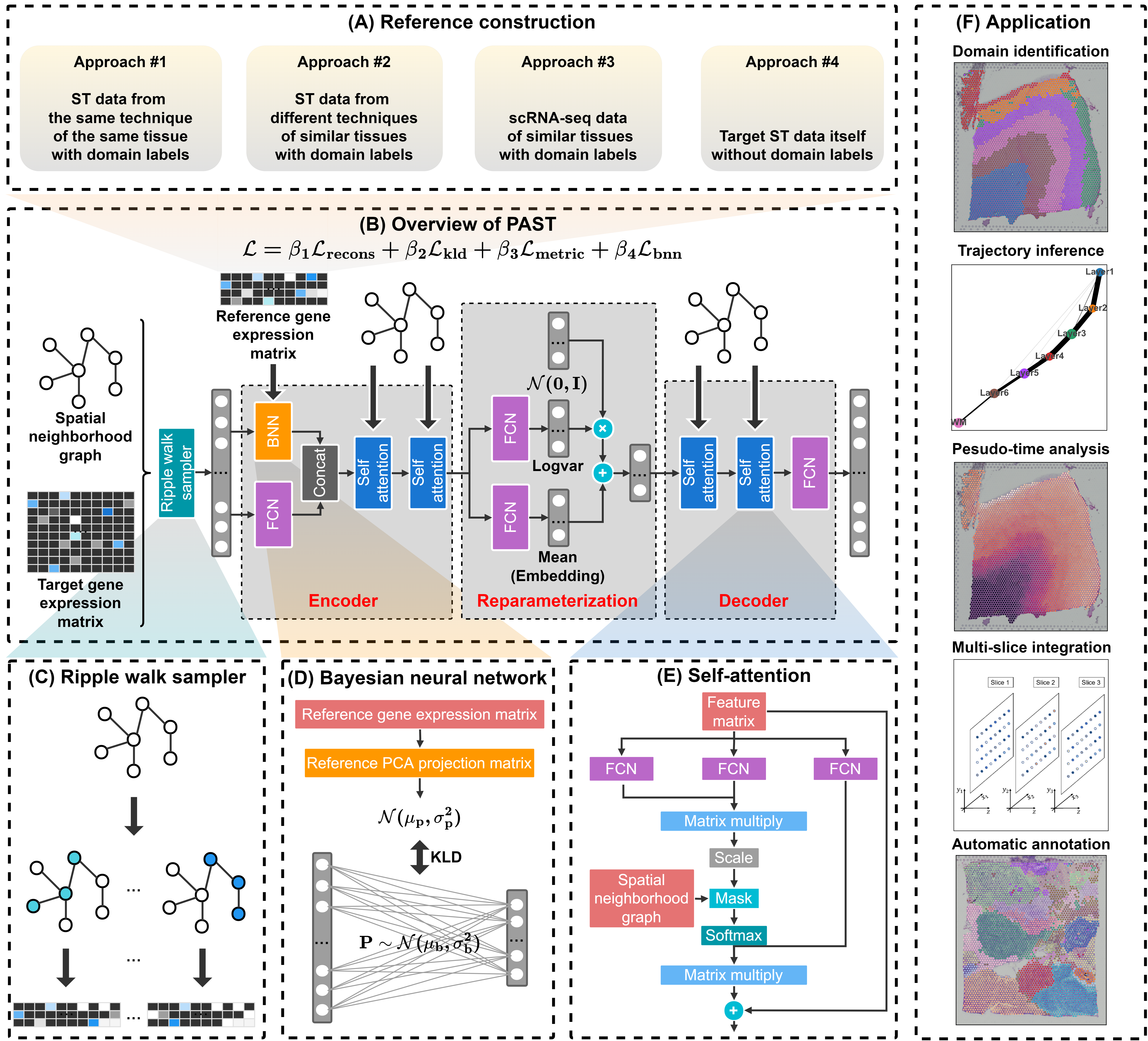
Rapid advances in spatial transcriptomics (ST) have revolutionized the interrogation of spatial heterogeneity and increase the demand for comprehensive methods to effectively characterize spatial domains. As a prerequisite for ST data analysis, spatial domain characterization is a crucial step for downstream analyses and biological implications. Here we propose PAST, a variational graph convolutional auto-encoder for ST, which effectively integrates prior information via a Bayesian neural network, captures spatial patterns via a self-attention mechanism, and enables scalable application via a ripple walk sampler strategy. Through comprehensive experiments on datasets generated by different technologies, we demonstrated that PAST could effectively characterize spatial domains and facilitate various downstream analyses, including ST visualization, spatial trajectory inference and pseudo-time analysis. Besides, we also highlight the advantages of PAST for multi-slice joint embedding and automatic annotation of spatial domains in newly sequenced ST data. Compared with existing methods, PAST is featured as the first ST method which integrates reference data to analyze ST data and we anticipate that PAST will open up new horizons for researchers to decipher ST data with customized reference data, which dramatically expands the applicability of ST technology.
Citation¶
Zhen Li, Xiaoyang Chen, Xuegong Zhang, Rui Jiang, and Shengquan Chen. “Latent feature extraction with a prior-based self-attention framework for spatial transcriptomics.” Genome Research (2023). doi:10.1101/gr.277891.123.

